ESM Fold
ESM Fold is a revolutionary protein structure prediction tool based on ultra-large language models, capable of directly inferring the complete three-dimensional structure from a protein's primary sequence with atomic-level accuracy. The model utilizes 15 billion parameters and learns from atomic-resolution images of protein structures, significantly improving the resolution of predictions and achieving an order-of-magnitude acceleration in protein structure prediction, marking a new chapter in the field.
The standout feature of ESM Fold is its combination of efficiency and high precision. It can complete large-scale predictions at ultra-fast speeds, accurately modeling over 225 million protein sequences and making structural inferences for more than 617 million metagenomic protein sequences, thereby constructing the ESM metagenomic map. Compared to traditional structure prediction methods relying on physical modeling or neural networks, ESM Fold achieves significant breakthroughs in computational efficiency and predictive capability, drastically reducing the demand for computational resources.
Additionally, ESM Fold shows tremendous potential in various application scenarios. Its efficient prediction capabilities make large-scale protein structure studies feasible, providing strong support for fundamental biological research, disease-related protein target discovery, drug design, and protein engineering. By precisely predicting the relationship between protein sequences and their three-dimensional structures, ESM Fold has advanced scientists' understanding of the molecular mechanisms of life and offers a novel solution for exploring protein sequences in unexplored metagenomes.
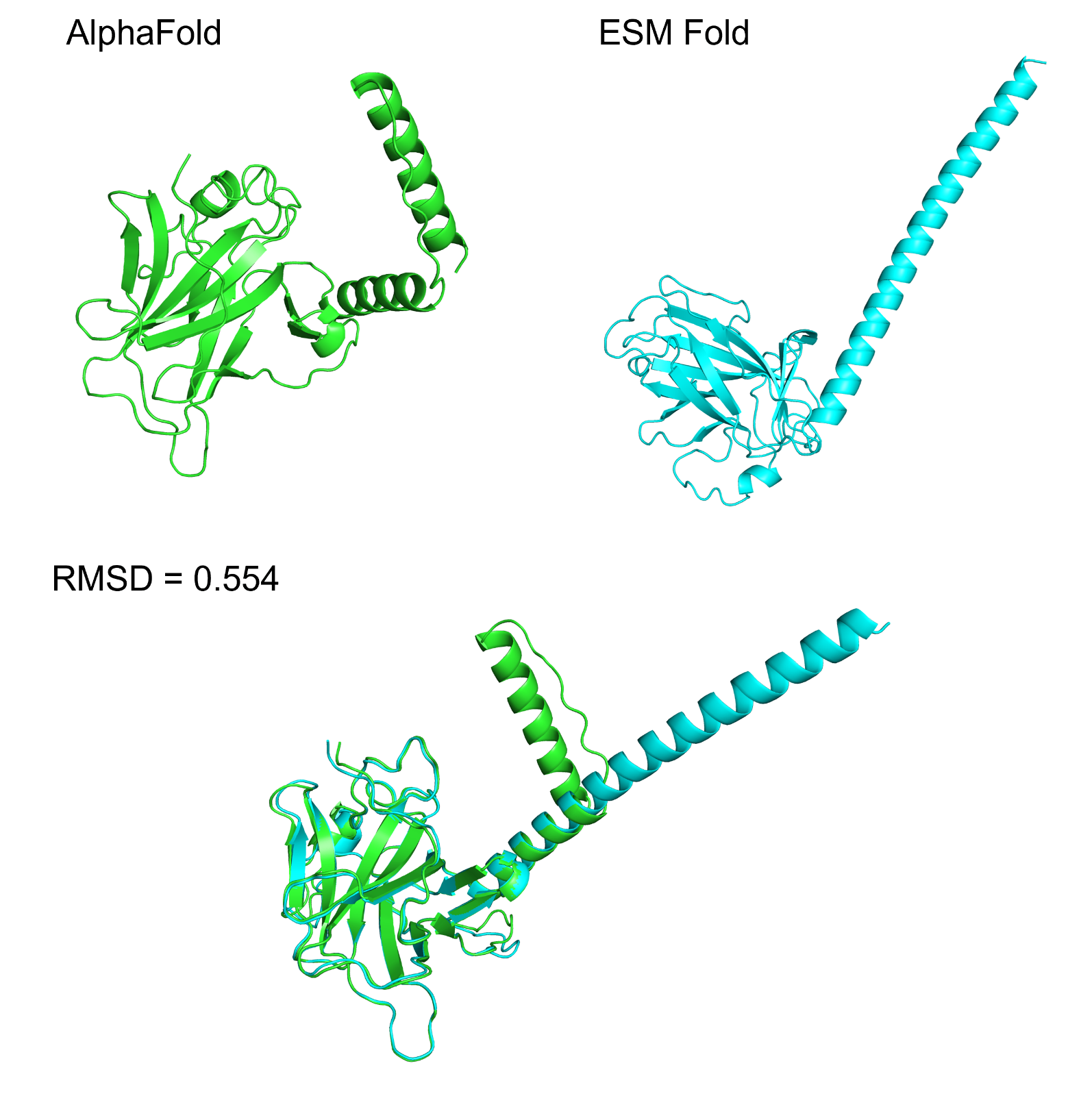
Figure 1: ESM Fold prediction of the protein model, showing minimal difference from the AlphaFold prediction model (RMSD=0.554).