ProteinMPNN
ProteinMPNN is a deep learning model specifically developed for protein sequence design, aimed at generating high-quality amino acid sequences based on a given protein structure. It has shown exceptional performance in the fields of protein engineering and directed evolution, providing powerful tools for designing novel proteins with specific functions.
Traditional protein-directed evolution requires repeated mutations and screenings to optimize protein function, which is time-consuming and inefficient. In contrast, ProteinMPNN can rapidly generate sequences that highly match the target structure using deep learning, significantly accelerating the protein design process. The model analyzes the structural features of the protein, predicts, and optimizes the arrangement of residues to design stable proteins with the desired functions.
ProteinMPNN excels in protein function optimization, binding affinity enhancement, and stability improvement, and is suitable for various fields such as enzyme engineering, antibody design, and synthetic biology. By combining with experimental directed evolution, ProteinMPNN offers more efficient and precise protein design solutions, driving advancements in protein engineering technology.
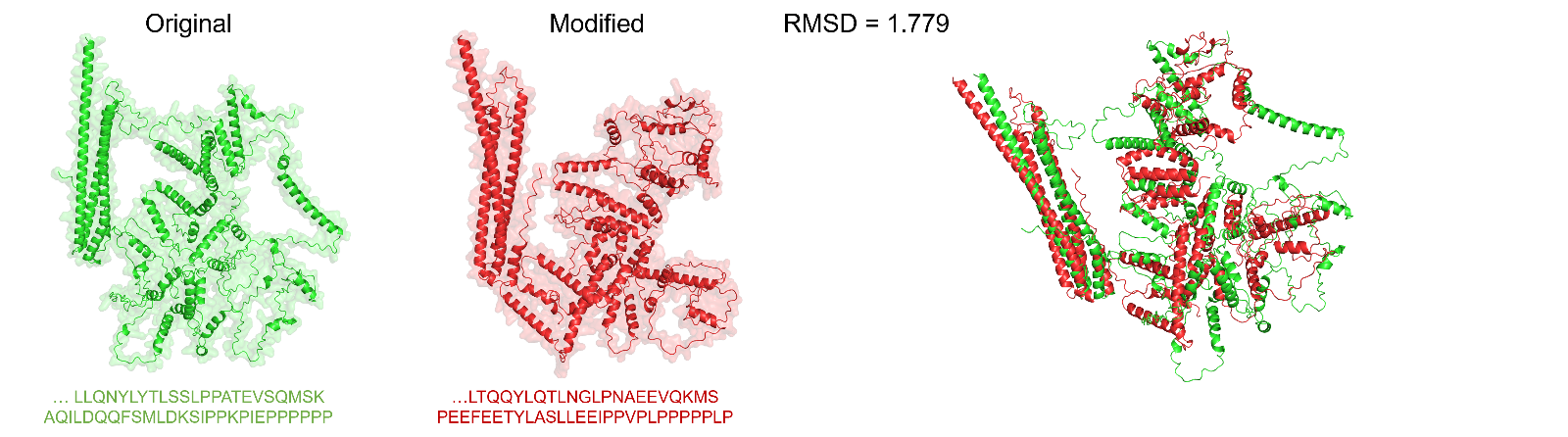
Figure 1: After modification with ProteinMPNN, the protein sequence undergoes significant changes, while the protein conformation shows minimal difference (RMSD=1.779).