Compound-protein docking
Release Date:2024-12-25
Publisher: Hefei Kejing
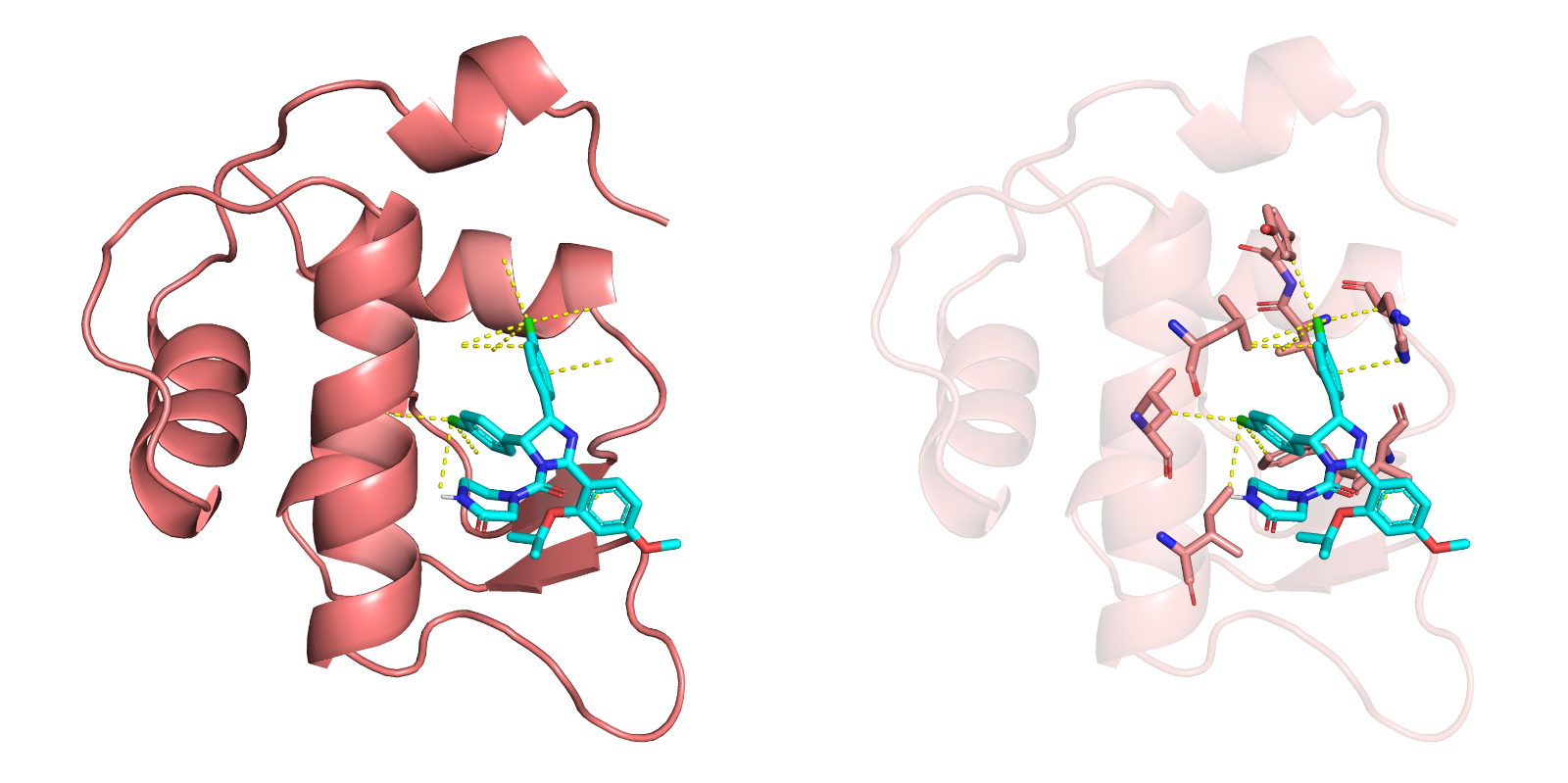
Compound-protein docking is used to predict the optimal binding mode between small molecules (such as drug candidates) and target proteins, providing critical support for drug discovery and design.
AutoDock Vina: Based on a free energy scoring function, AutoDock Vina accurately predicts the binding sites of compounds with proteins, making it an efficient and reliable tool for small molecule docking.
NeuralPlexer: Utilizing graph neural network technology, NeuralPlexer rapidly evaluates interactions between compounds and proteins, enhancing docking accuracy through deep learning.
DiffDock: By generating probability distributions of docking conformations and incorporating confidence analysis, DiffDock identifies the optimal binding mode between compounds and proteins, offering more comprehensive results for drug screening.
By integrating these three tools, we provide comprehensive predictions of compound-protein binding conformations, evaluate binding affinity, and perform confidence analysis on docking results, ensuring reliability and accuracy in the predictions.